-
Individual Report
review on May 31, 2018
by Rebecca Fishwick

At a Glance
Summary
I thought that the information provided by Gene Heritage was reliable, well-referenced and generous considering the low price. My Individual Report offered insight into some of my physical traits and their origins in a way that was clear and easy to understand. Had any of my parents or grandparents had their DNA analyzed, I would have been interested to see how our results compared, and from which of them I had inherited my different traits.
Full Review
Gene Heritage is an online service based in New York, USA, that allows people to upload their raw genetic data to provide family genetics analysis.
The company’s Technical Director, E. Castedo Ellerman, previously developed the open source software Genomology, which can perform DNA analysis across three generations of family members. Gene Heritage uses that same analysis software, presenting it in a way that is both understandable and visually attractive.
Their Creative Director, Joseph Silver, has been a designer for television networks and advertising agencies since 1994, and leads the design direction of Gene Heritage’s reports.
Product Expectations
The Gene Heritage website was simple and straightforward. They offered three services: an ‘Individual Report’, a ‘Parent-Child Report’, and a ‘Grandchild Report’. The Parent-Child Report was an analysis of both parents and their child, which would reveal which parent had passed which genes on to their child. Similarly, the Grandchild Report would reveal what genes had been passed on from each grandparent.
Since neither my parents nor my grandparents have their DNA analyzed, I opted for the Individual Report. I read that this would reveal the interesting variations in my genes, identify their ancient origins, and show me how they influence traits like eye color, and senses such as taste and smell.
I headed to the FAQs section to find out more, where I discovered that they accept genetic data from Ancestry DNA, 23andMe, Family Tree DNA, MyHeritage, Living DNA, National Geographic, and Genes for Good. They also provided instructions for downloading data from some providers.
I could also view a list of the different traits their tests covered, which included eye color, earwax type, armpit odor, lactose intolerance, “Asian Flush” (the “flush” response to alcohol), and taste and smell sensitivity. I learned that they did not report on genes that were of clinical importance.
I read that their reports could tell me how old my genes are: whether my genetic variations have been around for tens of thousands of years, or if they are the result of more recent mutations in Europe, Asia, Eurasia, or Africa.
I saw that I would be able to download my report, and use a “Share” feature to share my reports online if I wished. I could request for my DNA data to be deleted at any time.
Ordering Experience
In order to use Gene Heritage’s services, I would have to sign up for an account. Firstly, I had only to enter my email address. I saw that by clicking “Continue”, I was agreeing to the Terms of Service, and so I had a look at what these were.
Here, I found that by providing my email address, I was signing up for email marketing from Gene Heritage, which I could unsubscribe from (my email address would not be sold to third parties without my consent).
Additionally, by sharing my DNA data with Gene Heritage, I was releasing them from “any and all claims, demands, or suits in connection with the results of [my] Gene Heritage report, including, without limitation, errors, omissions, emotional distress, or economic loss”. I couldn’t imagine what economic loss I could incur from reading the report, or how learning that my eye color had originated in Eurasia – for instance – could cause emotional distress. They also emphasized that their reports were not paternity tests, and shouldn’t be used to “confirm or deny paternity relationships”.
It was clear the reports were intended for “educational purposes only”, and not for health care or legal purposes. They recommended speaking to a doctor or genetic counselor for any health concerns.
I read that while they tried to only report on genes that have a “strong influence on traits”, or to “clearly state when a gene only has a moderate to minor influence”, I should understand that there are other influences that determine the traits I have, and so there may be errors or inaccuracies.
By uploading my data to Gene Heritage, I was granting them permission to store my DNA data on their servers. They reserved the right to delete my data along with any reports generated within a week of my payment being submitted. Again, I saw I could request deletion of my data at any time.
I also looked at their Privacy Statement, where I read that Gene Heritage would not sell my DNA data or identifying information to third parties. For the purposes of safety and security, I wouldn’t be able to download my raw data file once it had been uploaded to the site (though presumably I would already have a copy). They also stated that they used cookies on the site.
Having looked at their policies, I was happy to continue.
Shortly after submitting my email, I was sent a private sign-in link, which would expire in seven days. Clicking the link, I was asked to enter a name for the person whose DNA data I was uploading (this didn’t have to be a full name). I could then upload a DNA file from my computer. The upload took approximately one minute.
The Individual Report had been put in my shopping cart (though I hadn’t made the purchase yet). I had the option of adding a parent or child, and I chose not to do so. I was then taken to my shopping cart. Clicking “Pay Now” took me to a PayPal link, where I could pay with a PayPal account, or with a credit or debit card.
Once I’d completed my payment, I was able to view my report.
The Results
In the “Your Reports” section, I was able to view or download my report. Since I’d read that my genetic data and my report could be deleted within a week, I decided to download a copy before viewing it online.
Results Section: Eye Color
The first result I had was for eye color. The gene they looked at was OCA2, of which I had inherited one “Light” allele, and one “Dark” allele. My allele pair determined which version of the gene I had (shown below).
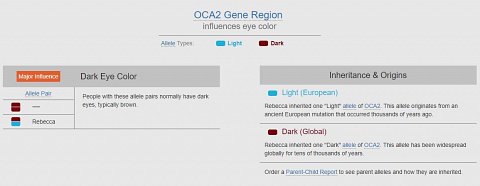
My eye color alleles.
Had I uploaded genetic information from one or both of my parents, I could have learned which parent gave me which allele (though for this sort of trait, it can be obvious just by looking at them!).
There was also expandable information about the ancestral origins of each allele, and about the OCA2 gene. I learned that the OCA2 gene produces melanin, a pigment affecting eye color, and so was a major influence on determining eye color. People with more melanin have darker eyes, whereas people with less melanin have lighter eyes.
I read that typically, African and Asian populations tend to have alleles for darker eyes, while alleles for lighter eyes are more common in European populations. One study showed that the mutation for lighter eyes occurred in Europe 6000 to 10,000 years ago.
I discovered that the OCA2 gene is not the only gene involved with determining eye color (TYRP1, ASIP, and ALC42A5 also contribute), but it is the most influential, and has some influence on determining skin and hair color too.
There was also a reference page for this gene, and pages for the other genes included in my results. The reference pages included links to articles about each gene, and information about the alleles involved in determining traits.
Results Section: Armpit Odor and Earwax
My next result covered armpit odor and earwax type, which – interestingly – were each covered by the same gene. Apparently, I’d inherited one copy of the “dry” allele and one copy of the dominant “wet” allele, meaning that I had wet earwax (shown below).

My earwax and odor types.
I honestly can’t say I’d ever noticed the texture of my earwax. And I also discovered – a little insultingly, perhaps – that since I had one copy of the “wet” allele, I was also slightly predisposed to having more armpit odor, though this was only given as a “Minor Influence”.
According to the information, the “dry” allele (of which I had one copy) had originated in Northeast Asia about 40,000 years ago, before spreading to other parts of the continent. The “wet” allele was most common among African and European populations.
I also read that earwax type and armpit odor are both produced by specialized skin glands, and that the ABCC11 gene is responsible for encoding a protein involved in apocrine secretion in those glands, which is why the same gene influences both.
Results Section: Taste Sensitivity
My results covered two genes that affect sensitivity to bitter tastes: the TAS2R38 gene and the TAS2R31 gene. The TAS2R38 gene affected taste sensitivity to PTC and PROP (two bitter-tasting chemicals), and similar bitter chemicals in vegetables.
I had inherited two copies of the “Sensitive” allele, and so I was more likely to perceive PTC, PROP, and cruciferous vegetables like broccoli, Brussel sprouts, cauliflower and cabbage as bitter.
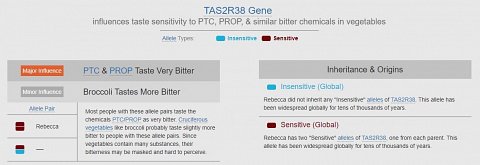
My variation of the TAS2R38 gene, affecting bitter tastes.
The TAS2R38 gene encodes instructions for building one of the types of bitterness taste receptors in the tongue’s taste buds. I read that the ability to detect bitterness may have helped our ancestors avoid toxic foods, though both the “Sensitive” and “Insensitive” alleles have been widespread for tens of thousands of years.
The TAS2R31 gene affected my bitterness sensitivity to saccharin, an artificial sweetener used in drinks, sweets, and medicines. I found I have one copy of the “Sensitive” allele, and one of the “Insensitive” allele, both of which have been widespread globally for a long time.
I read that this gene also influences sensitivity to caffeine and quinine (which is found in tonic water, and also used to treat malaria), and a chemical found in artichokes.
Results Section: Smell Sensitivity
The first of the genes covering smell sensitivity was OR5A1, which affects sensitivity to β-ionone: an organic compound found in violets and rose oil. I had both copies of the “Insensitive” allele, meaning that I was less sensitive to β-ionone, and therefore to floral fragrances. Both the “Sensitive” and “Insensitive” alleles were widespread globally (shown below).

My variation of the OR5A1 gene, affecting sensitivity to floral scents.
Another gene related to smell perception was OR2J3, which affects sensitivity to leaf alcohol: a grassy-scented plant substance. I had one copy of the “Sensitive” allele and one copy of the “Insensitive” allele, meaning I was less sensitive to grassy smells than some.
I read that the more sensitive allele had been around for tens of thousands of years and is widespread globally, while the more insensitive allele originated in Africa thousands of years ago, and spread into Eurasia. It is carried by around half of all Africans, and only an eighth of Eurasians.
The third gene related to smell sensitivity was OR2M7, which influences sensitivity to the pungent odorants in “asparagusic acid waste”, i.e. the pungent smell in urine after eating asparagus. I found I had both pairs of the “Insensitive” allele. Both alleles had been widespread globally for many thousands of years.
There was another gene affecting smell sensitivity, though my variation of it wasn’t included in my genetic data. This was the OR7D4 gene, which affects how individuals perceive androstenone, a mammalian pheromone.
Results Section: Alcohol and Lactose Tolerance
There were also genes influencing how the body tolerates alcohol and lactose. The ALDH2 gene affected how the body processes alcohol, and whether a person gets a flush response from consuming alcohol. I discovered that I had both copies of the “Off” allele, meaning that I don’t get any facial flush from alcohol.
This allele is widespread globally, whereas the “On” allele originated in Asia thousands of years ago, and is widespread among East Asian populations.
I read that alcohol flush is due to how the “On” allele interferes with the body’s ability to break down acetaldehyde, a toxic by-product of alcohol, which causes blood vessels to dilate and the skin to turn red.
I discovered that lactose tolerance was affected by the LCT gene region, including both the LCT gene that encodes lactase (the enzyme that helps to digest lactose, a sugar occurring in milk and dairy products) and the MCM6 gene, which regulates the production of lactase.
My version of the LCT gene had one “Off” and one “On” allele, meaning that I digest lactose better than people with both copies of the “Off” allele. The “Off” allele had been widespread globally for many thousands of years, while the “On” allele originated in Europe thousands of years ago.
Results Section: Sprinting Performance
The last gene included in the report was the ACTN3 gene, which has a minor influence on sprint performance. I found I had one copy of the “On” allele and one of the “Off” allele, meaning that I had some predisposition to faster sprinting ability (shown below).

My variation of the ACTN3 gene, to do with sprinting ability.
I read that although Olympic sprinters tend to have both copies of the “On” allele, there are many other factors that influence sprint ability, including environmental, diet and training factors, and other genes.
The “Off” allele, I read, was the result of a mutation in the ACTN3 gene that occurred in Eurasia thousands of years ago, while the “On” allele had been widespread across the globe for tens of thousands of years. I learned that the ACTN3 gene is responsible for encoding the Alpha-actinin protein, which affects the composition of skeletal muscle, and so influences sprint ability.
Summary
I thought that the information provided by Gene Heritage was reliable, well-referenced and generous considering the low price. My Individual Report offered insight into some of my physical traits and their origins in a way that was clear and easy to understand. Had any of my parents or grandparents had their DNA analyzed, I would have been interested to see how our results compared, and from which of them I had inherited my different traits.