-
Y Elite Ancestry Test for Men
review on 21 January 2016
by Craig Macpherson

At a Glance
Summary
I’m delighted that I undertook Y chromosome sequencing with Full Genomes. It’s been amazing to uncover my Finnish ancestry and allowed us to expand our family tree in a new direction. Full Genomes replied to all my questions promptly and were very sensitive to the fact that I’m not an expert.
This test is definitely not for the faint-hearted, there’s a very steep learning curve and once you start, there’s a mountain of research you can read through, dozens of projects to join, and dozens of additional analyses you can undertake. You’re not going to get any fancy user-friendly reports, but if you’re detail-orientated, researching your paternal ancestry can be very compelling.
I’d highly recommend this test to anyone who’s enthusiastic about their paternal ancestry. It’s been fun to get involved with the genetic genealogy community, and to help the team dedicated to mapping the paternal lineage of my haplogroup.
Full Review
I’ve taken many tests to analyse my Y chromosome and trace my paternal lineage, but always as part of a package of tests e.g. 23andMe. I know that Full Genomes are experts when it comes to Y chromosome sequencing for tracing paternal ancestry, and as sequencing provides data magnitudes greater than chip analysis (the type of analysis used by 23andMe, Family Tree DNA, Ancestry.com etc.), I couldn’t wait to try their ‘Y Elite 2.0 Sequencing’ test, to see what more I could learn…
Product Expectations
The Full Genomes site said they’d sequence my Y chromosome using 250 base pair reads at 50x coverage. In terms of the base pair reads, I didn’t really understand why this was significant. I know our DNA is 2.8 billion base pairs long, but the length of the reads seemed inconsequential to me. In terms of the ‘50x coverage’, I was aware that this indicates the accuracy of the sequencing. I’ve read that 30x coverage is the minimum required for accurate results, so I was impressed that I’d get 50x with the Y Elite 2.0.
I also read that Full Genomes would analyse my SNPs (Single Nucleotide Polymorphisms aka genetic variants, aka mutations), and my STRs (Short Tandem Repeats aka genetic patterns) – both SNPs and STRs can be used to link your Y chromosome to shared paternal ancestors.
When the analysis was complete, I’d discover my paternal haplogroup which designates the specific group of ancestors my Y chromosome links me to. I’d also receive my ‘Private SNPs’ which correspond to recent mutations not yet observed, access to the Full Genomes database to find living relatives, and I’d be shown where I’m placed on the Y chromosome phylogenetic tree – the Y chromosome phylogenetic tree shows how Y chromosomes have mutated and branched over time.
Ordering Experience
The Full Genomes site was easy to use and purchasing was straightforward, I had to pay a $50 shipping fee as I was ordering from the UK. Three days after placing my order I received an email to say the sample kit had been shipped, and four days later I received the kit.
The kit contained an invoice for $18 for two spit kits and shipping which was odd, but after contacting Full Genomes, they reassured me that I didn’t have to pay this. In order to send the sample back to Full Genomes, I had to pay another $5 for postage. It was a shame that I didn’t receive confirmation that my sample had been received.
Several weeks after dispatching my sample, I received an email inviting me to set up an online account with Full Genomes where I’d access my results.
The Results
A few weeks later I received an email to say that my results were ready. I logged into my account and was invited to download my ‘Interpretation Results’. The Interpretation Results contained several files with instructions, and these were split by Sequencing lab analysis files and Full Genomes analysis files.
The Sequencing lab analysis files contained the raw data which were generated when my Y chromosome sequence was run. The files focussed on four types of genetic variant: CNVs, INDELs, SNPs and SVs. Although instructions were included for each type, they were very complex, and as Full Genomes had undertaken a separate analysis to the sequencing lab, I decided to focus on their analysis instead.
Results Section: Full Genomes analysis files
The Full Genomes analysis folder contained two files, a variantCompare and haplogroupCompare file. These are reports that can be opened using Excel and that show my high-reliability ‘private variants’. These reports also allowed me to compare my results to other anonymous Full Genomes customers, identify recent mutations, and place my variants on the Y chromosome phylogenetic tree.
It should be said that the variantCompare and haplogroupCompare reports contained thousands of rows, and I had no idea how to extract any meaningful information from the data. When I asked Full Genomes, they said the haplogroupCompare report showed my paternal haplogroup was ‘S4744+’ (also known as ‘CTS11603’), but when I looked in the report myself, this designation appeared alongside hundreds of others in the ‘Named variant status’ column (shown below), so I wasn’t really sure why S4744+ was more significant than the others.
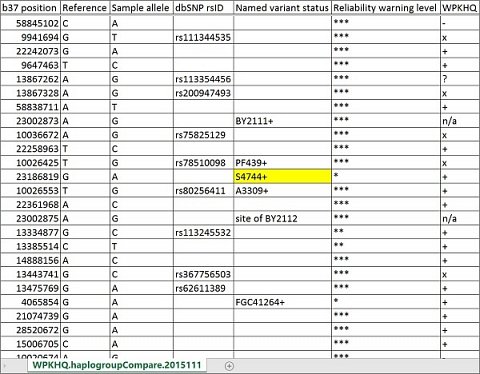
My S4744+ paternal haplogroup designation in the haplogroupCompare report.
Results Section: The Y chromosome phylogenetic trees
As well as telling me that my paternal haplogroup was S4744+, Full Genomes also told me what my equivalent haplogroups were – this is because different organisations use different Y chromosome phylogenetic trees.
In the ‘Composite tree’ https://sites.google.com/site/compositeytree/i I was told my haplogroup appears as: ‘I1a1b4 L300/S241’ and ‘CTS11603/S4744’ and ‘23186819 G->A’ (shown below):

My paternal haplogroup in the composite tree.
In the ISOGG – International Society of Genetic Genealogy – tree http://www.isogg.org/tree/ISOGG_HapgrpI.html I was told my haplogroup also appears as ‘I1a1b4 L300/S241’ (shown below):
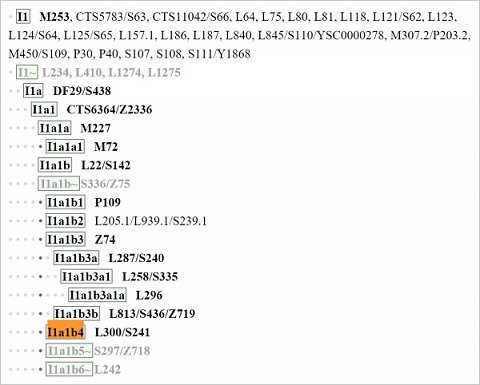
My paternal haplogroup in the ISOGG tree.
Results Section: Those I share a paternal ancestor with
Full Genomes revealed that my Y chromosome was closely linked to eight other records on their database (shown below). It’s likely that I share a paternal ancestor with these people, and possible that one or several of these men are my direct paternal ancestor. This was fascinating! A previous test with Britains DNA revealed that my recent (within the last thousand years) paternal ancestors are from Scandinavia, so it was great to see this partially corroborated.
• Thomas Thompson, b. 1731, d. 1796
• Elias Myllykangas, circa 1760, Kuivasmäki, Petäjävesi, Finland
• Isaac b.1778, Ireland – and his descendant – Thomas Banks Wright, b. 1825
• Mikael Mikaelinpoika, b. 1560, d. 1607, Rauma, Finland
• August Pässinkivi, circa 1856, Hunsala, Loppi, Finland
• Mikko Juhonpoika, b. 1765, Taivassalo, Finland
• Johan Hammern, circa 1640
It was amazing to see that records indicate many generations of my paternal ancestors were Finnish. I discussed it with my Dad who’d never discussed his paternal lineage with his own father. We were both impressed with the analysis and pleased that we have a better idea of where our male ancestors come from.
After discussing with Full Genomes, they advised me to email Steve Trangsrud who’s one of the experts for the ‘I1’ haplogroup (which encompasses the ‘I1a1b4’ haplogroup) and they directed me to this page on the Family Tree DNA site for more information: https://www.familytreedna.com/public/yDNA_I1
I learned that the I1 haplogroup is estimated to be four to five thousand years old and confirmed by the SNP ‘M253’. Apparently many I1 members trace their ancestry to Scandinavia, and others find their roots in the British Isles, Germany, Central Europe, Iberia and Russia.
Results Section: Why Full Genomes and Britains DNA indicated different haplogroups
Interestingly, my test with Britains DNA confirmed my paternal haplogroup as S4744+, but put me in subgroup ‘I-S142’ which is linked to Scandinavian ancestry. After following up with Full Genomes, I learned that ‘S241’ (the paternal haplogroup I’m in according to the Composite and ISOGG trees) is a subgroup of I-S142. Full Genomes sent me the following extract of the ISOGG tree to help explain:
• • I1a DF29/S438
• • • I1a1 CTS6364/Z2336
• • • • I1a1a M227
• • • • • I1a1a1 M72
• • • • I1a1b L22/S142 <————————————my haplogroup according to Britains DNA
• • • • • I1a1b1 P109
• • • • • I1a1b2 L205.1/L939.1/S239.1
• • • • • I1a1b3 Z74
• • • • • • I1a1b3a L287/S240
• • • • • • • I1a1b3a1 L258/S335
• • • • • • • • I1a1b3a1a L296
• • • • • • I1a1b3b L813/S436/Z719
• • • • • I1a1b4 L300/S241 <————————————my haplogroup according to Full Genomes – one level deeper
It’s clear that working out your position on the Y chromosome phylogenetic tree is complicated, especially given the different types of taxonomy and the number of levels in each. I wished there’d been an easier way to see my haplogroup on the trees as part of the results!
Other reports in the Full Genomes analysis folder let me look at my data in different ways: gtype reports showed my results in relation to tens of thousands of known SNPs, yKnot reports showed where my haplogroup fits on the ISOGG Y chromosome phylogenetic tree, and Y-STR reports showed the unique Y chromosome STRs I possess.
Summary
I’m delighted that I undertook Y chromosome sequencing with Full Genomes. It’s been amazing to uncover my Finnish ancestry and allowed us to expand our family tree in a new direction. Full Genomes replied to all my questions promptly and were very sensitive to the fact that I’m not an expert.
This test is definitely not for the faint-hearted, there’s a very steep learning curve and once you start, there’s a mountain of research you can read through, dozens of projects to join, and dozens of additional analyses you can undertake. You’re not going to get any fancy user-friendly reports, but if you’re detail-orientated, researching your paternal ancestry can be very compelling.
I’d highly recommend this test to anyone who’s enthusiastic about their paternal ancestry. It’s been fun to get involved with the genetic genealogy community, and to help the team dedicated to mapping the paternal lineage of my haplogroup.


